As treatment for operable colon cancer evolves from adjuvant to neoadjuvant chemotherapy, there has been an increased need to scrutinize the performance of CT for risk stratification -- particularly in distinguishing high-risk colon cancer with adverse features from low-risk colon cancer without such features, according to Prof. Vicky Goh, chair of clinical cancer imaging at King's College London.
In a commentary published on 21 July by European Radiology, she contended that there is still a need for improvement in the use of CT imaging for risk stratification, and therefore room for other complementary approaches.
Goh, who will become interim editor in chief of Radiology in August 2025, cited a previous systematic review and meta-analysis as a metric for the current iteration of this use of CT imaging: CT staging showed a sensitivity and specificity of 90% (95% confidence interval [CI]: 83, 95%) and 69% (95% CI: 62, 75%), respectively, for detecting T3-T4 and T1-T2 cancers, and 67% (95% CI: 46, 83%) for node positivity.
The variations would be related to factors such as differences in reader experience, imaging acquisition and reconstruction, and suboptimal imaging planes, she added.
 Prof. Vicky Goh is soon to become interim editor-in-chief of Radiology. Photo courtesy of KCL.
Prof. Vicky Goh is soon to become interim editor-in-chief of Radiology. Photo courtesy of KCL.
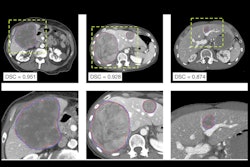
The use of radiogenomics can contribute to the accuracy of CT imaging here, according to Goh. She cited a retrospective two-center study by Caruso et al in which a contrast-enhanced CT clinicoradiomic model was used to predict pathologically defined high-risk colon cancer in a cohort of 300 patients, the results of which were promising.
The final clinicoradiomic model had a sensitivity of 86%, but a lower specificity of 48% for predicting ≥ pT3 colon cancer (area under the receiver operating characteristic curve [AUC], 0.74). However, in a subset of 118 patients who had also undergone RNA and DNA sequencing, a separate CT radiogenomic model that included more variables had a sensitivity of 88% and specificity of 63% for predicting ≥ pT3 colon cancer (AUC, 0.84).
Goh added that the staging performance was not as good as that of two radiologists (AUC, 0.82; sensitivity 94%, specificity 68%) for pathologically defined high-risk cancer (≥ pT3) in the entire sample; however, this was expected, and the performance of the model exceeded that of a nonexpert radiologist (AUC, 0.58; sensitivity 68%, specificity 47%).
While these results suggest that the model could be useful, Goh noted that this study is typical in showing the challenges of radiomic/radiogenomic studies: smaller sample size, potential for overfitting, and a lack of external validation. In addition, the authors could provide more information so that the models could be tested independently, such as code or image preprocessing.
Moreover, the specificity remained low, and while variables were added, “it is not clear from the data presented how much the radiomic and genomic features added to the performance of multivariable models with these established variables alone," Goh observed.
However, Goh suggested that while radiogenomics is not quite ready to make up areas where CT imaging may still be lacking in risk stratification, Caruso et al’s findings suggest it to be a fruitful area of inquiry.
Read the article here.