Researchers from the Institute for AI in Medicine (IKIM) at University Hospital Essen, Germany, have developed an open-source AI tool that combines algorithms for multiple imaging deidentification steps.
Furthermore, the technique produces results for DICOM MRI, CT, and whole slide images, as well as MRI twix raw data, comparable to those of state-of-the-art algorithms but with significantly reduced computational time, according to findings published in European Radiology on 7 June by Moritz Rempe, Lukas Heine, and colleagues.
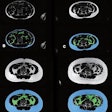
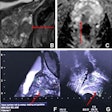
 Comparison of the defacing results of different defacing algorithms. While the result of pydeface and the proposed algorithm are similar, pydeface additionally cuts off the shoulder region of the scan, while taking 260 times longer on average than the proposed algorithm. The face shown is from the publicly available Synthstrip dataset.Courtesy Rempe, Heine et al, European Radiology
Comparison of the defacing results of different defacing algorithms. While the result of pydeface and the proposed algorithm are similar, pydeface additionally cuts off the shoulder region of the scan, while taking 260 times longer on average than the proposed algorithm. The face shown is from the publicly available Synthstrip dataset.Courtesy Rempe, Heine et al, European Radiology
Medical imaging data used in research often includes sensitive protected health information (PHI) and personal identifiable information (PII), both of which are regulated under rigorous legal frameworks such as the General Data Protection Regulation (GDPR) or the Health Insurance Portability and Accountability Act (HIPAA).
Furthermore, advances in imaging technology -- and the ever-increasing sophistication of malicious actors -- require that data be deidentified before use. Balancing the need to safeguard patient privacy in compliance with regulations while facilitating data exchange presents challenges, especially given the complexity and variety of imaging data.
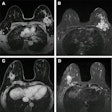
Medical image deidentification consists of multiple separate tasks: metadata anonymization, in which all personal meta information (e.g., name, sex, date of birth, diagnosis) in the data header is removed; defacing, in which the face is removed from the medical image while the rest of the image remains; skull-stripping, which leaves the brain as the only output; text removal; and whole slide image (WSI) deidentification. Each of these tasks required different datasets for the tool to be trained on.
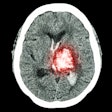
 Skull-stripping results of the proposed algorithm. Shown are examples from the Synthstrip test dataset, including T1, magnetic resonance angiography (MRA), proton density (PD) and infant T1 scans. The proposed methods produce sound results, but might struggle with parts of infant brains.Courtesy Rempe, Heine et al, European Radiology
Skull-stripping results of the proposed algorithm. Shown are examples from the Synthstrip test dataset, including T1, magnetic resonance angiography (MRA), proton density (PD) and infant T1 scans. The proposed methods produce sound results, but might struggle with parts of infant brains.Courtesy Rempe, Heine et al, European Radiology
The researchers designed their tool as a Python3 Command Line Interface (CLI) application as well as a standalone Docker container. The input data is initially read (specific to the data type), and then different deidentification tasks are performed. Metadata removal or pixel data cleaning, including skull-stripping, defacing, or text removal, can be performed for all common medical imaging data types, they noted.
A "defacing score" was set to compare the technique with algorithms already in use, calculated by the number of scans, which the face recognition model cannot classify as faces anymore. Additionally, the authors tracked the computation time per volume for each algorithm; the computation time is an important factor for real-time algorithms.
The tool’s algorithms for defacing and skull-stripping performed similarly to the compared state-of-the-art algorithms, but at a significantly reduced computation time: the computation time per volume was reduced by up to 260 times, to an average time of 0.88 ± 0.15 seconds, compared with 233.57 ± 59.87 seconds needed by one of the compared algorithms, pydeface. The authors noted that the tool performed fastest with GPU support, but also attained better times on CPU-only devices. For skull-stripping, the proposed tool outperformed the algorithm BET, while achieving results similar to those of Synthstrip and HD-BET and with reduced computational time as well.
The methods included for the skull-stripping dataset were T1-weighted MR angiography, proton density (PD), and infant brain scans. The authors noted that the algorithm had some difficulty in skull-stripping infant brain scans, leaving some residue in the output.
The proposed text removal algorithm had a deidentification score of 83.59% on ultrasound images. The algorithm performed well for metadata deidentification, with all specified metadata correctly modified according to the specified DICOM profile. The metadata deidentification was verified by a human tester on a dataset comprised of 410 files (DICOM and DICOM-WSI files), they added.
The new algorithm has some limitations. For one, the text removal algorithm takes out all text, which can include text that should remain for future tasks. This, the researchers suggested, can be modified in future iterations.
In addition, they prioritized speed over accuracy gains in cutting the extensive preprocessing that other algorithms used: "This speedup is particularly important in clinical practice, when large amounts of data have to be processed on a daily basis."
Read the European Radiology article here.