Artificial intelligence (AI) algorithms are capable of segmenting cerebral white-matter hyperintensities of presumed vascular origin on brain MR images as well as or better than human observers, enabling more reliable quantification of these lesions, according to the results of a recent AI developer challenge.
A multi-institutional and multinational team of researchers organized a scientific challenge for AI software developers and, importantly, found that a number of the submitted deep-learning models could produce equivalent performance on images acquired at different institutions and on different scanner types.
"Using these methods in clinical practice can help to more precisely quantify the amount and distribution of [white-matter hyperintensity] lesions," said lead author Hugo Kuijf, PhD, from University Medical Center Utrecht in the Netherlands. "Since the location of [white-matter hyperintensities] might be more relevant than the actual volume, the resulting segmentations can provide these measurements for use in lesion-symptom mapping studies."
Analyzing white-matter hyperintensities
 Hugo Kuijf, PhD, of University Medical Center Utrecht. Image courtesy of Rozanne Land.
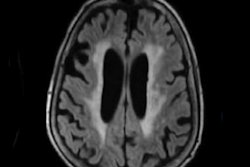
Hugo Kuijf, PhD, of University Medical Center Utrecht. Image courtesy of Rozanne Land.Small-vessel disease plays a key role in stroke, dementia, and aging. One of its main consequences is white-matter hyperintensities, which can be visualized on MRI. The current clinical practice of visually rating the presence and severity of white-matter hyperintensities has considerable limitations, however, according to Kuijf. Although automatic white-matter hyperintensity quantification methods do exist, it's hard to compare them objectively because all are evaluated using a different ground truth and different criteria.
"A further challenge of automatic methods is the deployment in a new institute that has different scanners and protocols," he told AuntMinnieEurope.com. "Methods may not always generalize well and, thus, the performance of existing methods on new data is unknown."
The researchers shared the results of their WMH Segmentation Challenge in an article published online on 19 March in IEEE Transactions on Medical Imaging.
In the challenge, Kuijf and colleagues sought to address these issues and to perform a standardized assessment of various automatic methods using data from multiple sites and on images acquired from scanners that the algorithms hadn't seen before. They launched their WMH Segmentation Challenge at the International Conference on Medical Image Computing and Computer Assisted Intervention in 2017 (MICCAI 2017). To participate, teams first had to register on the challenge's website; they could then download 60 sets of T1 and fluid-attenuated inversion-recovery (FLAIR) brain MR images with manual annotations of white-matter hyperintensities from three different institutes and MRI scanners.
Twenty participants submitted their algorithms for evaluation on a test set of 110 brain MR images from five different scanners -- including two that weren't included in the training data.
The researchers found that many of the deep-learning methods were capable of generalizing to unseen data from unseen scanners. They also discovered that the top-ranking algorithms shared a number of methodological properties: the use of neural networks, dropout layers during training, some form of hard negative learning, and ensemble methods, Kuijf said.
"The top-ranking methods perform above the interobserver variation, so these methods might assist or replace individual human observers," he said.
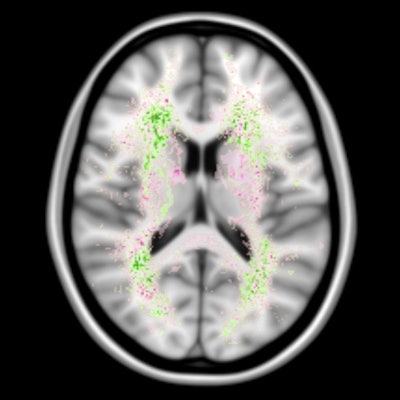
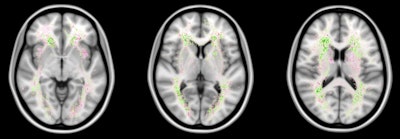 False negatives are the most common source of error for both humans and computers. The images show a spatial distribution of the false-negative white-matter hyperintensity segmentations of a human observer and the top 4 ranking methods in the WMH Segmentation Challenge. The colors indicate the differences. Green means that the top 4 methods had fewer false negatives, while pink means that the human observer had fewer false negatives. Overall, the top 4 AI methods had fewer false negatives than a human observer, but the AI methods made more mistakes at locations where lesions are rarely found. Images courtesy of Hugo Kuijf, PhD.
False negatives are the most common source of error for both humans and computers. The images show a spatial distribution of the false-negative white-matter hyperintensity segmentations of a human observer and the top 4 ranking methods in the WMH Segmentation Challenge. The colors indicate the differences. Green means that the top 4 methods had fewer false negatives, while pink means that the human observer had fewer false negatives. Overall, the top 4 AI methods had fewer false negatives than a human observer, but the AI methods made more mistakes at locations where lesions are rarely found. Images courtesy of Hugo Kuijf, PhD.
The most common source of error for the algorithms was missing white-matter hyperintensity lesions that were very small. This will be a target for future developments, Kuijf said.
An open challenge
Meanwhile, the challenge remains open for new and updated submissions. Since the challenge's inception at MICCAI 2017, seven new or updated methods have been submitted and the online leaderboard is kept up to date, he said.
"Researchers are encouraged to use the data to develop methods for conference or journal publications and have their methods evaluated independently on the test set," Kuijf said. "One new submission and four updated methods have entered the top 10 of the challenge, considerably changing the leaderboard. Ensemble methods still perform very well and a second method using multidimensional gated recurrent units has entered the top 3."
Kuijf also said that most participants have agreed to make their algorithms available as open access on the challenge's Docker Hub, enabling others to replicate the results.